Investigating Protein Dynamics, Structure and Function using Mass Spectrometry
How proteins work is very closely related to how they are shaped and how they move. That’s right, proteins don't just sit there looking pretty (as it appears in an X-ray crystallography structure). Their shape is constantly fluctuating but these motions are not random, they are in fact, function critical with defined start and end points. A crystal structure shows the most stable conformation but in many cases the weakly populated unstable conformation is the source of biological activity!
In our lab we use Hydrogen Deuterium Exchange (HDX) and Ion mobility (IM) in conjunction with mass spectrometry (MS) to study protein dynamics and interactions. Mass Spectrometry, as the name suggests, is a way of detecting molecules according to their mass. We use Electrospray Ionization (ESI) MS which has the additional benefit that we can distinguish - in a very rough way - different protein shapes by looking at how charged they become in the electrospray process.
We use a technique that Dr. Derek Wilson introduced in his PhD with Dr. Lars Konermann that facilitates Time-Resolved ElectroSpray Ionization (TRESI) MS. This allows us to use our highly sensitive and selective mass spectrometers to study rapid processes (like protein motions, which occur mostly on the millisecond timescale).
Over the past several years, many systems have been studied using the previously described techniques. You will find a brief summary of these systems here, as well as a list of students with experience in each field. Feel free to get in contact with either Dr. Wilson (dkwilson@yorku.ca), or any of the members listed here if you need more information.
Enzymes:
Enzymes are a class of protein molecules that catalyze chemical reactions. We rely on them to make otherwise ridiculously slow reactions occur at a pace that is suitable for life as we know it. Enzymes are usually highly specific, which is to say that they often target a specific molecule (the substrate) whilst leaving other ones alone. Over the past few years we have worked with several interesting enzymes in our lab including: β-lactamases (e.g. TEM-1 and OXA-58), Glutathione S-Transferase (GST) and Yeast Alcohol Dehydrogenase.
β-lactamases are bacterial enzymes that are responsible for antibiotic resistance as they cleave β-lactam antibiotics such as ampicillin and penicillin rendering them useless. Currently, our lab is looking at the allosteric hotspots of the enzyme while it binds to a variety of antibiotic substrates. GST is a family of detoxification enzymes found in both prokaryotes and eukaryotes that catalyzes the conjugation of GSH (an antioxidant). GST is a super family of proteins consisting of 6 classes (zeta, pi, alpha, omega, mu and theta). Alcohol dehydrogenase (ADH) is a group of enzymes that catalyze the conversion of alcohols to aldehydes or ketones and reduce NAD+ to NADH.
Key Publications: Rob et al. (2013), Liuni et al. (2013), Knox et al. (2018)
Intrinsically Disordered Proteins (IDPs):
Intrinsically disordered proteins and domains have long represented a challenge to structural biologists. Disordered proteins often fail to crystalize and typically do not provide sufficient electron density to calculate an X-ray crystallographic structure when they do. NMR can provide significant insights in some cases, however, the lack of a well-defined native structure tends to broaden-out signals or cause low dispersion in NMR spectra, making conventional structural NMR analyses exceedingly challenging (and often impossible). One of the most exciting features of TRESI-MS/HDX is the ability to characterize residual structure in intrinsically disordered proteins. A central protein of interest that has been studied in our lab is the human Tau protein, which causes the formation of neurofibrillary tangles and is linked to neurodegeneration in Alzheimer’s disease when it becomes misfolded. Currently, we are studying the changes in dynamics of Tau upon ligand binding, to help us better understand its mechanism of interaction, and paving the way to find therapeutic agents. Another IDP of interest is amyloid beta, a main culprit in Alzheimer's disease. The dynamics of this protein is being studied by our lab using molecular modeling and molecular dynamics simulations, with MOE and Gromacs software.
Student Contact: Banafsheh Mehrazma
Key Publications: Zhu et al. (2015), Zhu et al. (2016)
Protein-Protein Interactions:
Protein molecules often interact with other proteins (e.g. Antibody-Antigen complexes) and also self associate to form dimers, tetramers and oligomers (e.g. K122-4 pilin from P. aeruginosa).
Antibodies are produced in the immune system in the response to an antigen which is usually a protein. In our lab we perform epitope mapping, which is a method of identifying the antibody binding sites on the antigen of interest. An example of an antigen we have worked with is myoglobin, an iron and oxygen binding protein found in muscle cells.
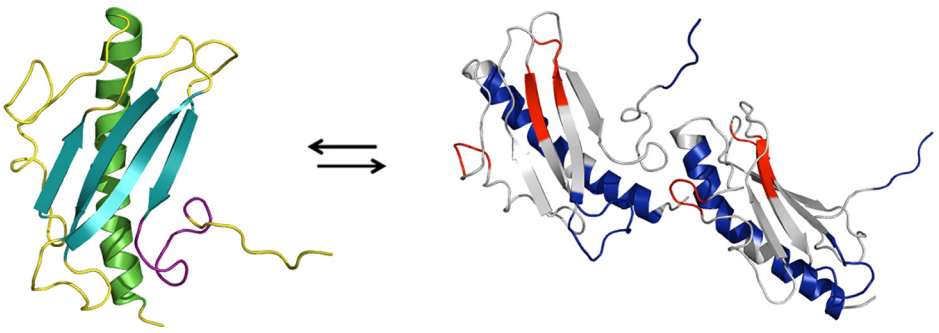
Protein oligomerization is involved in the formation of bacterial pili structures responsible for various infections. For example, the formation of the type IV pilus (constructed from type IV pilin eg. K122). K122 has been shown to enter a pre-oligomerization stage in which it enters a monomer-dimer equilibrium. This structural change that occurs from monomer to dimer has been previously studied in our lab. This is a collaborative research project with Dr. Gerald Audette's lab at York University.
Key Publications: Lento et al. (2015), Deng et al. (2017)
Protein-Ligand Interactions:
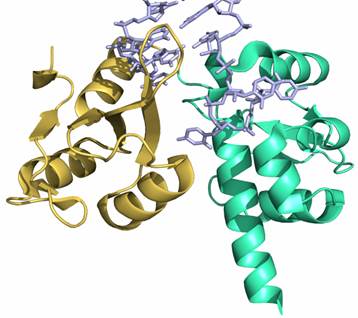
Proteins also interact with RNA and DNA ligands in a variety of different ways. An important class of these molecules are RNA chaperones that are involved in the correct processing, folding and maturation of RNA ligands. In our lab we study structural changes that occur in RNA-chaperones such as the human La protein (hLa), when bound to RNA ligands. This is a collaborative research project with Dr. Mark Bayfield's lab at York University.
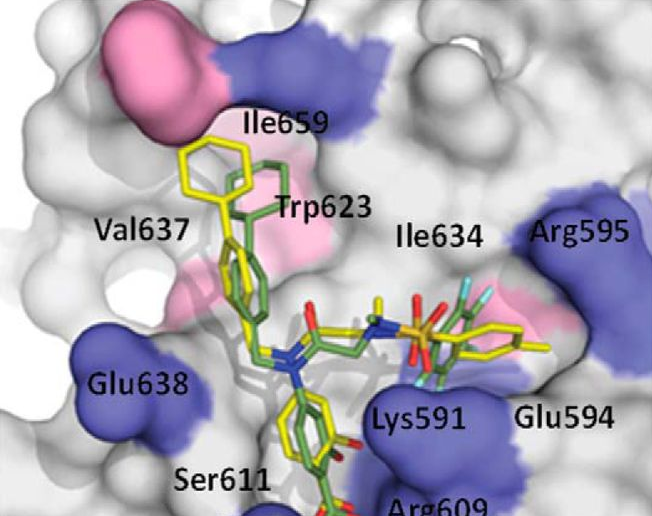 In the past, studies were conducted to probe local and global pertubations upon interaction of salicylic acid-based inhibitors with the Src Homology 2 (SH2) domain in the Signal Transducer and Activator of Transcription 3 (STAT3) protein.
In the past, studies were conducted to probe local and global pertubations upon interaction of salicylic acid-based inhibitors with the Src Homology 2 (SH2) domain in the Signal Transducer and Activator of Transcription 3 (STAT3) protein.
Currently, we are working in collaboration with the University of Chicago to study the human antiapoptotic proteins Bcl-2 and Mcl-1, whose overexpression in cancers enables cells to evade death when they’re past their “expiry date” and accumulate mutations. The process of apoptosis, or programmed cell death, is essential in maintaining homeostasis with respect to cell turnover. However, cell death (or lack of it) is also a major player in the development of cancers and chemotherapy resistance. Despite similarities in tertiary structure, Bcl-2 and Mcl-1 are able to bind distinct BH3 domains of proapoptotic counterparts (Bid, Noxa, etc.). Time-resolved HDX-MS will be employed to examine how structural dynamics part-take in the ability of Bcl-2 and Mcl-1 to accomodate various ligands.
Student Contact: Esther Wolf
Key Publications: Resetca et al. (2014), Brown et al. (2016)
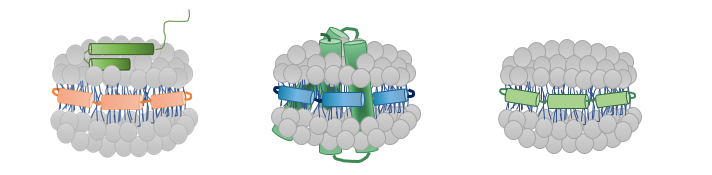
Protein-Membrane Interactions Using Nanodiscs:
Each field has its own “Mount Everest”, and the modern-day drug industry is no exception. There is a great challenge in studying the interactions between membrane proteins and the phospholipid lipid bilayer of the cell/vesicles.
Recent advances on membrane protein research in vitro involves the use of phospholipid nanodiscs as the native-like bilayer environment. Nanodiscs enable structural–functional investigations of membrane proteins which has been limited thus far. Nanodiscs are non-covalent structures of a phospholipid bilayer and membrane scaffold proteins (MSPs) which resembles the structure of a sandwich – lipid bilayer polar heads on top and bottom interact with the solvent, the hydrophobic tails interact with each other and MSP and are in the middle of the nanodisc. Our group sees the great potential in looking at the dynamics of protein-membrane interactions using nanodiscs coupled to TRESI-HDX technology.